oxLink™ and sxLink™ Photocrosslinking Technologies
| Technique Note TN202101 |
| oxLink™ and sxLink™ Products |
Dynamic biomolecular interactions are responsible for many cellular functions. Heterobifunctional photo-crosslinking reagents are time-tested tools for studying these interactions. Typically, these reagents are small organic compounds containing one chemical group for anchoring to the biomolecule of interest and another photoactivatable group for covalently crosslinking the interacting partner(s). At CellMosaic, we have developed two types of proprietary photo-crosslinking reagents: the oxLink™ and sxLink™ reagents (Figure 1). These reagents combine the highly efficient carbene-generating phenyldiazirine group with convenient reversible chemical crosslinking and the super hydrophilic AqT™ linker.
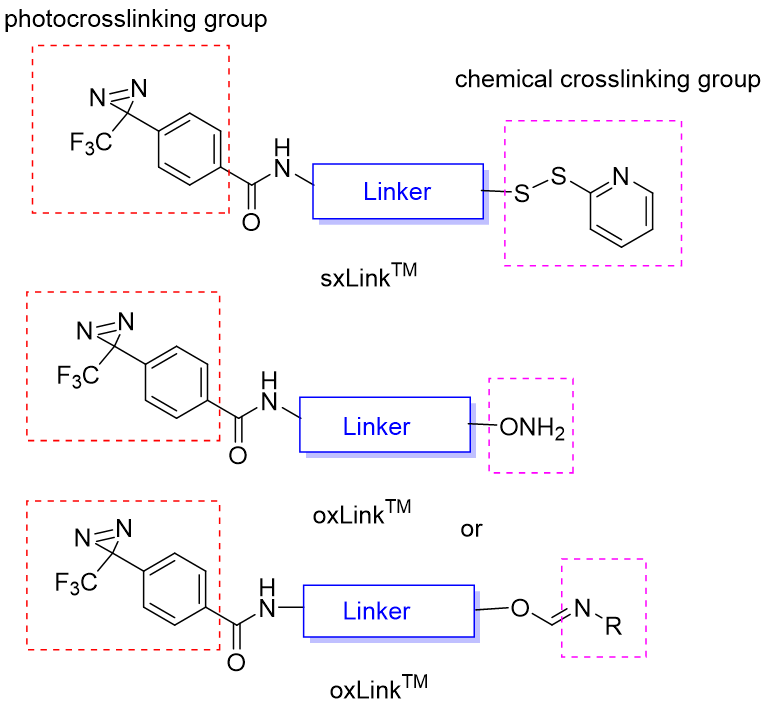
Figure 1. General chemical structures of oxLink™ and sxLink™ reagents. A typical regent contains (i) a phenyldiazirine photoactive group that photo-crosslinks the interacting biomolecular partner; (ii) a chemical crosslinking group (thiol or aminooxy) capable of forming a reversible linkage after crosslinking to a biomolecule or a preformed releasable linkage (disulfide bond or oxime) with a chemical crosslinking group; and (iii) a flexible linker that tethers these two functional groups. The releasable linkage can be used for downstream purification, enrichment of crosslinked products, and identification of crosslinking sites, in combination with advanced mass spectrum analysis.
Photo-crosslinking Group
The properties of the photo-crosslinking reagents are crucial for obtaining reliable crosslinking data. First, the reagent must be stable under the experimental conditions. Second, the photolabile group should possess appropriate photolysis characteristics, meaning, the photolysis conditions should not interfere with or alter the function of the biomolecules involved. Third, the photo-generated reactive intermediates should be highly reactive and should not rearrange into less reactive intermediates.
CellMosaic’s oxLink™ and sxLink™ reagents utilize the highly efficient carbene-generating 3-trifluoromethyl-3-phenyldiazirine as the photo-crosslinking group. 3-Trifluoromethyl-3-aryldiazirines photolyze at longer UV wavelength (~360 nm), minimizing photodamage to biomolecules. The generated carbene inserts into C−H bonds within picoseconds. Additionally, the electron-withdrawing trifuoromethyl group stabilizes the intermediate diazo-isomer, preventing the formation of side products under normal labeling conditions.
Chemical Crosslinking Group
CellMosaic’s sxLink™ reagent contains a sulfhydryl-specific pyridyl thiol group, allowing attachment to specific cysteines or free thiols on a biomolecule. If the biomolecule lacks a free thiol group, sxLink™ can also be synthesized with a preformed disulfide bond linked to a chemical crosslinking group, such as carboxylic acid, enabling labeling of native biomolecules.
The oxLink™ reagent contains an aminooxy group for chemical crosslinking of biomolecule with ketone or aldehyde groups. Additionally, oxLink™ can be synthesized with a preformed oxime bond linked to a chemical crosslinking group, such as carboxylic acid, for labeling native biomolecules. Aminooxy-ketone ligation is orthogonal to peptide chemistry and allows highly selective, solid phase-based separation of aminooxy-labeled crosslinked products. This selective purification method is especially useful for characterizing protein complexes in complex matrices, such as plasma, cellular membranes, and cell lysates, where free thiols may interfere with labeling and purification processes.
Linker
oxLink™ and sxLink™ can be linked via ethylene, ethylene glycol, or superhydrophilic AqT™ linker. AqT™ linkers are novel proprietary biomaterials invented at CellMosaic, chemically assembled from a class of natural and edible sugar alcohol (SA) compounds designed for specific properties. Since the 3-trifluoromethyl-3-phenyldiazirine group is highly hydrophobic, biomolecules labeled with a 3-trifluoromethyl-3 phenyldiazirine compound using traditional ethylene or ethylene glycol-type linkers tend to aggregate and destabilize the labeled protein.
The oxLink™ and sxLink™ reagents with AqT® linkers significantly increase water solubility, improve biocompatibility, and reduce non-specific hydrophobic interactions with other biomolecules. These AqT® based photo-crosslinking reagents also allow for high loading of phenyldiazirine groups, greatly enhancing the chances of successful photo-crosslinking.
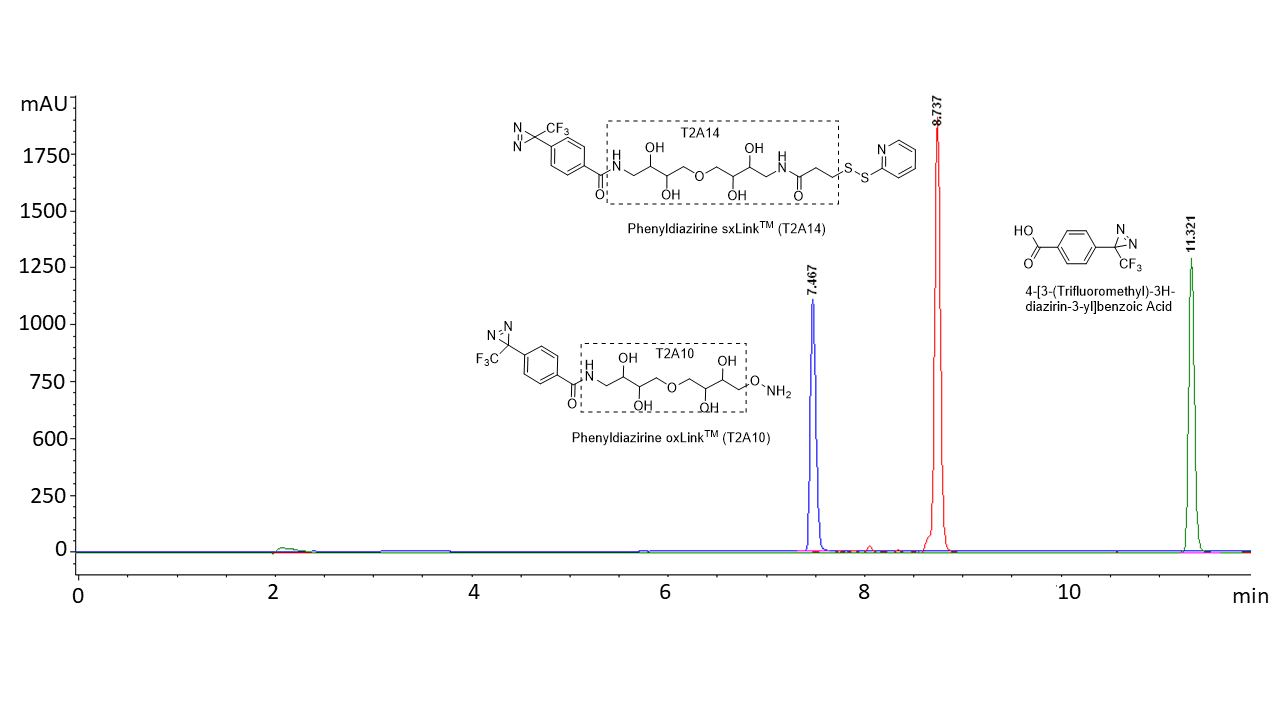
Figure 2. C18 HPLC analysis of 4-[3-(trifluoromethyl)-3H-diazirin-3-yl]benzoic Acid (green, not soluble in water), phenyldiazirine oxLink™ (T2A10) (blue, highly soluble in water: >27 mg/mL), and phenyldiazirine sxLink™ (T2A14) (red, moderately soluble in water: 2.2 mg/mL saturated solution). HPLC method: Buffer A: 0.1% TFA in water; Buffer B: 0.1% TFA in acetonitrile. Gradient: 5% to 95% Buffer B over 12 minutes.
Application and Workflow for oxLink™ and sxLink™
Figure 3 and 4 illustrate the workflow for using a reversible thiol or oxime linker to crosslink the interacting biomolecule partner, as well as to identify the crosslinking site or detect the interaction partner.
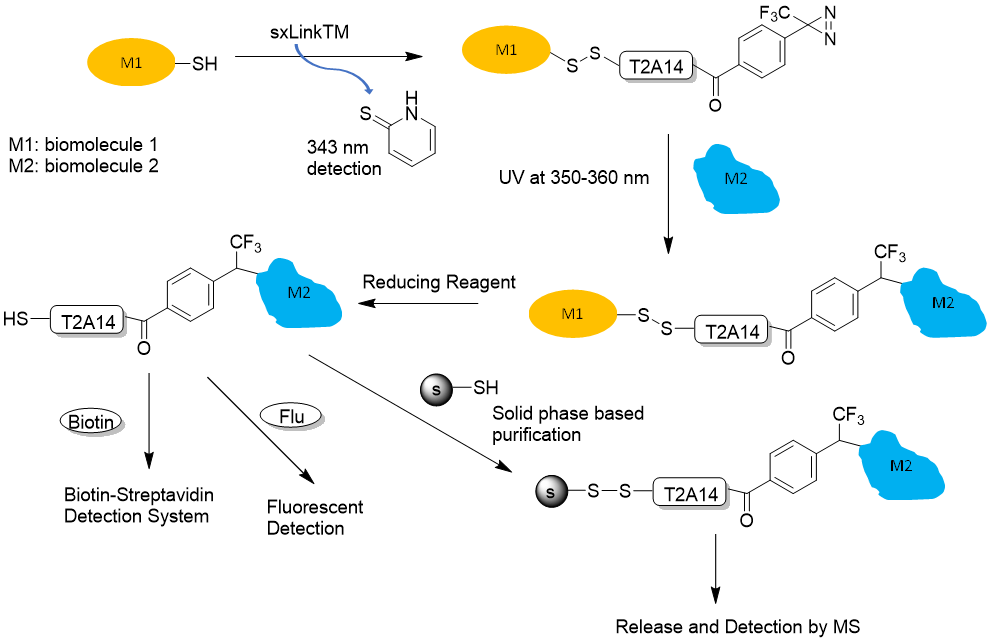
Figure 3. sxLink™ workflow using water-soluble phenyldiazirine sxLink™ (T2A14): First, sxLink™ is tethered to a target biomolecule of interest (M1) via a chemical disulfide crosslinker (thiol exchange). The progression of labeling can be monitored by UV analysis of the released chromophore, 2-thiolpyridone at 343 nm. The modified biomolecule (M1) is then reconstituted into its native complex with biomolecule M2. Next, the phenyldiazirine group is activated by UV irradiation, resulting in a crosslink with the neighboring biomolecule (M2). Finally, the disulfide bond between the two biomolecules is cleaved by a reducing reagent, revealing the biomolecule interaction through the transfer of the free thiol to M2.
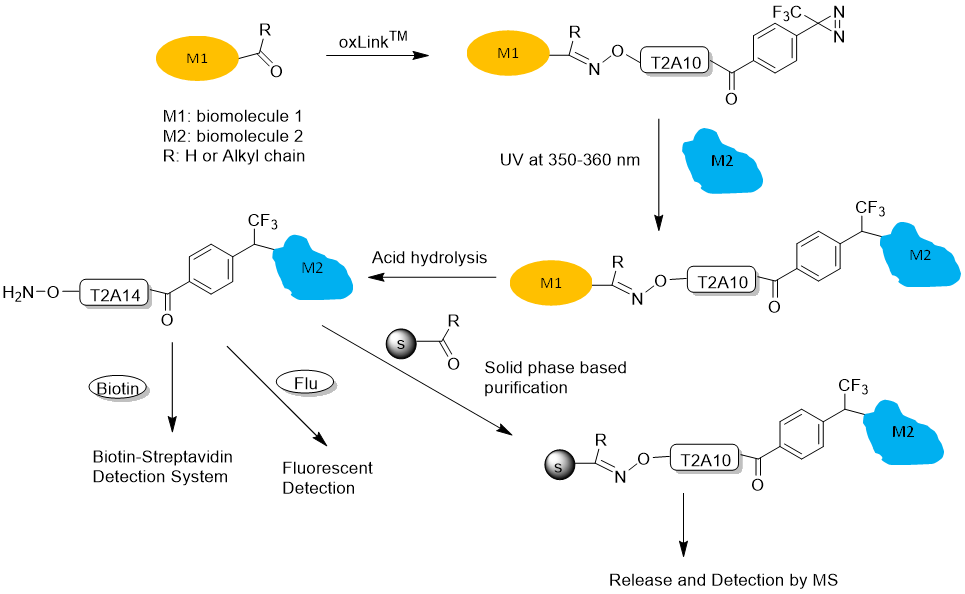
Figure 4. oxLink™ workflow using water-soluble phenyldiazirine oxLink™ (T2A10): First, oxLink™ is tethered to the target biomolecule of interest (M1), which has an artificially introduced ketone or aldehyde group, via the aminooxy functional group. The resulting oxime bond can be hydrolyzed, releasing the photo-crosslinked biomolecule (M2) with a free aminooxy group. M2 can then react with any aldehyde- or ketone-containing molecule, such as a solid phase, biotin, or fluorescent dye, for further purification, detection, and identification.
Patent and license restrictions
oxLink™ and sxLink™ crosslinking reagents are covered under U.S. patent numbers 8907079B2, 9511150B2, US14/965,699, and equivalent patents and patent applications in other countries, in the name of CellMosaic, Inc. The purchase of oxLink™ and sxLink™ products includes a limited license for internal research and development, but not for any commercial purposes. Commercial use includes, but is not limited to:
1. The sale, lease, license or other transfer of the material or any material derived or produced from it.
2. The sale, lease, license or other grant of rights to use this material or any material derived or produced from it.
3. The use of this material to perform services for a fee for third parties, including contract research and drug screening.
If you are interested in using oxLink™ and sxLink™ reagents for commercial purposes, please contact us for more information.
References
[1] Protein crosslinking reviews: a) Brunner, J. (1993) Annu. Rev. Biochem. 62, 485−514. b) Freedman, R. B. (1979) Trends Biochem. Sci. 193–197. c) Herrmann, J. M., Westermann, B., Neupert, W. (2001) Methods Cell Biol. 65, 217–230. d) Fancy, D. A. (2000) Curr. Opin. Chem. Biol. 4, 28–32. e) Fasold, H., Klappenberger, J., Meyer, C., Remold, H. (1971) Angew. Chem. Internat. Edit. 10, 795–801. d) Bayley, H. Photogenerated Reagents in Biochemistry and Molecular Biology, Vol. 12. Elsevier, Amsterdam, Neth, 1983.
[2]. Protein crosslinking reviews: a) Brunner, J. (1993) Annu. Rev. Biochem. 62, 485−514. b) Freedman, R. B. (1979) Trends Biochem. Sci. 193–197. c) Herrmann, J. M., Westermann, B., Neupert, W. (2001) Methods Cell Biol. 65, 217–230. d) Fancy, D. A. (2000) Curr. Opin. Chem. Biol. 4, 28–32. e) Fasold, H., Klappenberger, J., Meyer, C., Remold, H. (1971) Angew. Chem. Internat. Edit. 10, 795–801. d) Bayley, H. Photogenerated Reagents in Biochemistry and Molecular Biology, Vol. 12. Elsevier, Amsterdam, Neth, 1983.
[3]. Gritsan, N. P., Gudmundsdóttir, A. D., Tigelaar, D., Zhu, Z., Karney, W. L., Hadad, C. M. & Platz, M. S. (2001) J. Am. Chem. Soc. 123, 1951−1962.
[4]. a) Smith, R. A. G., Knowles, J. R, (1973) J. Am. Chem. Soc. 95, 5072−5073. b) Smith, R. A. G., Knowles, J. R, (1975) J. Chem. Soc. Perkin Trans. 2, 686−694.
[5]. Brunner, J., Senn, H. & Richards, F. M. (1980) J. Bio. Chem. 255, 3313−3318.
[6]. a) Nassal, M. (1983) Liebigs Ann. Chem. 1510−1523. b) Nassal, M. (1984) J. Am. Chem. Soc. 106, 7540−7545.
[7]. Marriott G, Ottl, J. (1998), Meth Enzymol 433, 155−175.
[8]. Reversible thiol linkage for studying rhodopsin and transducin interactions: a) Resek, J. F., Bhattacharya, S.& Khorana, H. G. (1993) J. Org. Chem. 58, 7598−7601. b) Resek, J. F., Farrens, D. & Khorana, H. G. (1994) Proc. Natl. Acad. Sci. USA 91, 7643−7647. c) Cai, K, Itoh, Y. & Khorana, H. G. (2001) Proc. Natl. Acad. Sci. USA 98, 4877−4882. d). Huang Y, Khorana HG. (2003) Mapping of Contact Sites in Interaction between Transducin and Light-Activated Rhodopsin. Presented at 17th Symposium of the Protein Society, July 26–30, Boston, Massachusetts.
[9]. Rose K, Zeng W, Regamey P, Chernushevich IV, Standing KG, Gaertner HF. (1996) Natural peptides as building blocks for the synthesis of large protein-like molecules with hydrazone and oxime linkages. Bioconjugate Chem 7:552-556.